In 1884, histones were discovered by Albrecht Kossel. The word “histone” is derived from the German word “histon”.
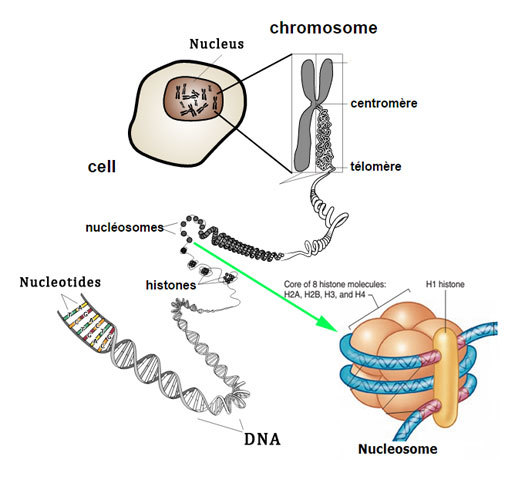
The nuclear DNA in all the eukaryotic cells is extremely compact through their connection with histones, special proteins that neutralise their electrostatic feature. These proteins are richly supplied with arginine and lysine seen in eukaryotic cells.
Thus, Histones are a special group of proteins seen in the nuclei of eukaryotic cells involved in the formation of chromatin and DNA folding. These are highly alkaline basic proteins and are positively charged.
Histones serve as spools for DNA that spiral into it, creating structural units referred to as nucleosomes, appearing as if beads on strings. These nucleosomes, in turn, are bound into 30-nanometer fibres, forming compacted chromatin. These histones restrict the DNA from getting twisted and tangled, thereby protecting it from any damage to the DNA.
Additionally, these histones are involved in DNA replication and the regulation of genes. In their absence in the chromosomes, the unwound DNA would be too lengthy. For instance, in every cell of humans, there are approximately 1.8 meters of DNA if it is stretched out completely, but when it is twisted about the histones, its length decreases to approximately 0.09 mm of the 30 nm diameter chromatin fibres.
Classes of Histones
There are two primary classes of Histones:
- Core histones – families included are H2A, H2B, H3, H4
- Linker histones – families included are – H1, H5
Two of each of these core histone proteins assemble to form an octameric nucleosome core particle, while 147 base pairs of DNA surround their substance.
The H1 linker histone proteins associate the nucleosomes at the start and end sites of DNA, hence securing DNA in place and helping in the formation of structures of higher orders.
Histone Octamer Definition – What is Histone Octamer?
Histone octamers are a complex of eight positively charged histone proteins that assist in DNA packaging. These are seen at the centre of the nucleosome core particle. Histones comprise two copies of each of the 4 core histone proteins – H2A, H2B, H3 and H4. This octamer gathers when a tetramer consisting of two copies of H3 and H4 composites with 2 dimers of H2A/H2B.
They assist in the nucleosome-formation. Negatively charged DNA surrounds the positively charged histone octamer to form nucleosomes.
Each of the histones has a C-terminal histone-fold and an N-terminal tail, both of which factors are involved in interactions with the DNA through a sequence of feeble interactions, even the salt bridges and the hydrogen bonds. Such association holds the histone octamer and DNA associated loosely, enabling both to completely separate and rearrange.
Histone Octamer Diagram
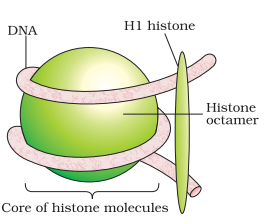
Histone Octamer Structure
The structure of the octameric histone core of nucleosomes was observed under X-ray crystallography to a 3.1 A resolution. This histone octamer is a tripartite assembly wherein a tetramer is centrally placed and flanked by dimers – H2A-H2B.
Their DNA is retained by ionic bonds. The linked histone HI associates with each nucleosome wherein the DNA enters and exits, thus drawing a row of nucleosomes towards it, forming a fibre of 10 nm. Nucleosomes in the chromatin appear as beads-on strings when seen under an electron microscope.
There is a complex outer surface that seems like a flat disk or a wedge. This disk indicates the planar projection of a proteinaceous superhelix (left-handed) with about 28 A pitch.
The core histones are 4 proteins referred to as H2A, H2B, H3 and H4 seen in equal parts in cells. All these four histone amino acid sequences comprise 20%-24% lysine and arginine. Protein size ranges between 11400 and 15400 Daltons, causing them to be comparatively smaller and still highly positively charged proteins. This high content of the charged (positive) amino acids enables them to be associated closely with the DNA (negatively charged).
The histone fold domains lead to the formation of heterodimers, which happens when the core histones engage with the quasi-symmetric heterodimer. The symmetry enables the heterodimer to be placed on itself by a 180-degree rotation around the axis of symmetry. Consequently, both ends of the histones which are involved in the binding of the DNA of the H3-H4 (crescent shape) are equivalent, still organizing the different DNA stretches. In a similar way, the H2A-H2B dimer folds too. In the first step of the formation of the nucleosome, the H32-H42 tetramer is enclosed with the DNA surrounding it. After this, both the H2A-H2B dimers are associated with the DNA- H32-H42 complex-forming nucleosome.
In addition to the histone-fold domains, each of the 4 core domains also comprises unstructured, flexible extensions referred to as histone tails. These are treated to a range of alterations such as lysine and arginine residues, acetylation, methylation of serine and phosphorylation.
Histone Octamer Function
Histones are a group of proteins associated with DNA in the nucleus and aid in condensing them into chromatin. This nuclear DNA is highly condensed and surrounds the histones in first order to be able to get accommodated in the nucleus and participate in the chromosomal formation.
Each histone octamer comprises two copies of histone proteins H2A, H2B, H3, and H4. Then the series of nucleosomes is enclosed in a spiral referred to as a solenoid, wherein the extra H1 histone proteins are linked with each nucleosome for the maintenance of the structure of the chromosome.
Apart from DNA compacting, these histone octamers are involved in the transcription of the DNA which it surrounds. These octamers interact with the DNA via both their N-terminal tails and core histone folds. These hold physically and chemically interact with the minor groove of the DNA.
Besides compacting the DNA, these octamers are involved in the DNA transcription that it surrounds.
Nucleosomes – Histone Octamer
The basic units of chromatin, nucleosomes, package and regulate the expression of the genomes in eukaryotes. These are extremely dynamic and remodelled using ATP-dependent remodelling factors.
Their structure comprises a DNA segment wrapped around the 8 histone proteins and seems as though a thread is wrapped around a spool. It consists of a little less than 2 rounds of DNA surrounding the histone octamers. Nucleosomes comprise histone octamers wrapped by 146 base pairs of DNA, surrounded in a superhelical arrangement.
Recommended Video:
Types of Nucleic acids and Structure of DNA | BIOLOGY | NEET | Concept of the Day
Frequently Asked Questions on Histone octamer
Are histones positively or negatively charged?
Histones are positively charged molecules, allowing tighter bonds to the negatively charged molecules of DNA.
In DNA tertiary structure, what is a histone octamer?
Histone octamers help in DNA packaging. Nucleosomes are formed when the negatively charged DNA surrounds the histone octamers. This DNA is held by ionic bonds. The linker histone H1 associates with each nucleosome wherein the DNA enters and leaves, drawing a series of nucleosomes towards forming 10 nm fibre. In the chromatin, the nucleosomes appear as beads-on string structures under an electron microscope.
This was a brief on histone octamer. For more on NEET, visit BYJU’S.
Comments