MeaningFunctionStructure and TypesProkaryotic DNA PolymeraseEukaryotic DNA PolymeraseMechanism of Action
DNA polymerases are a group of enzymes required for DNA synthesis. Arthur Kornberg purified and characterized DNA polymerase from E.coli for the first time. It is a single-chain polypeptide now known as DNA polymerase-I. Scientists have now found five DNA polymerases in E. coli.
Download Complete Chapter Notes of Molecuar Basis of Inheritance
Download Now
DNA Polymerase Definition
“DNA Polymerases are a group of enzymes that catalyse the synthesis of DNA during replication.”
The main function of DNA polymerases is to duplicate the DNA content of a cell during cell division. They do so by adding nucleotides at 3’-OH group of the growing DNA strand.
DNA Polymerase Function
Replication
The main function of the DNA polymerase is to synthesize DNA by the process of replication. It is an important process to maintain and transfer genetic information from one generation to another. DNA polymerase works in pairs, replicating two strands of DNA in tandem. They add deoxyribonucleotides at the 3′-OH group of the growing DNA strand. The DNA strand grows in 5’→3’ direction by their polymerisation activity. Adenine pairs with thymine and guanine pairs with cytosine. DNA polymerases cannot initiate the replication process and they need a primer to add to the nucleotides.
| Also check: NEET 2022 Answer Key PDF |
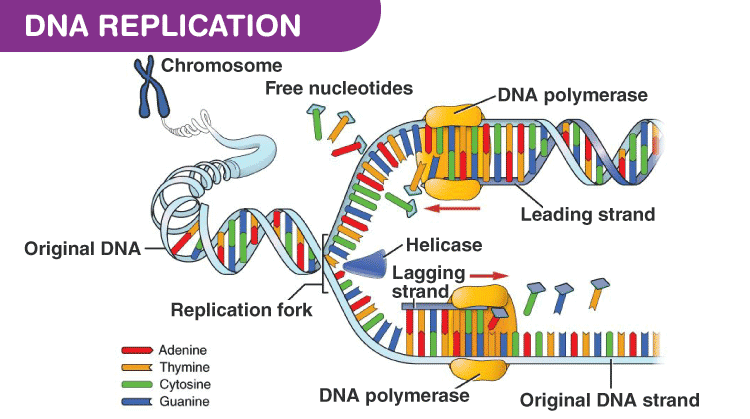 DNA polymerase III is the main enzyme responsible for replication in prokaryotes. In eukaryotes, DNA polymerase 𝝳 is the main enzyme for replication.
DNA polymerase III is the main enzyme responsible for replication in prokaryotes. In eukaryotes, DNA polymerase 𝝳 is the main enzyme for replication.
DNA polymerase I removes the RNA primer by 5’→3’ exonuclease activity and replaces the primer by its polymerase activity in the lagging strand.
Explore more: When does DNA copying occur?
Repair
The replication process is a humongous task and it is important to maintain the integrity of the genome. Apart from replication errors, DNA repair is the continuous process to rectify any errors in the genome due to DNA damage. There are various mechanisms by which DNA is repaired.
Proofreading
DNA replication is not perfect and there occurs an error after every 104 to 105 nucleotides added. Removing the incorrect nucleotide sequence or mismatched nucleotides from the newly synthesised strand is very important for the functionality of proteins, which can even lead to cancer. DNA polymerases remove incorrect pairs by exonuclease activity. They move one step back and remove the mismatched pair by 3’→5’ exonuclease activity. This is known as proofreading.
DNA polymerases are also involved in the post-replication DNA repair processes and also in translesion synthesis by which DNA polymerase copies unrepaired part of the DNA blocking the progression of replication.
Let’s learn in detail about different types of DNA polymerases and their functions.
Also see: DNA Ligase
DNA Polymerase Structure and Types
The structure of most of the DNA polymerases resembles a hand, which is holding active sites. The active site of the enzyme has two parts. At the insertion site, nucleotides are added. After adding, the newly formed base-pair migrates to the post-insertion site.
Prokaryotic DNA Polymerase Types and Function
There are five DNA polymerases identified in E.coli. All the DNA polymerases differ in structure, functions and rate of polymerization and processivity.
DNA Polymerase I is coded by polA gene. It is a single polypeptide and has a role in recombination and repair. It has both 5’→3’ and 3’→5’ exonuclease activity. DNA polymerase Ⅰ removes the RNA primer from lagging strand by 5’→3’ exonuclease activity and also fills the gap.
DNA Polymerase II is coded by polB gene. It is made up of 7 subunits. Its main role is in repair and also a backup of DNA polymerase III. It has 3’→5’ exonuclease activity.
DNA Polymerase III is the main enzyme for replication in E.coli. It is coded by polC gene. The polymerization and processivity rate is maximum in DNA polymerase III. It also has proofreading 3’→5’ exonuclease activity.
DNA polymerase III of E.coli is made up of a total of 13 subunits, which comprises 9 different types of subunits.
- It consists of two core domains made up of 𝜶, 𝟄, and 𝞱 subunits. It is attached to the 𝝲 complex or clamp-loading complex, which is made up of five subunits, 𝞽2𝝲𝝳𝝳’. Additional subunits 𝟀 and 𝟁 are attached to the clamp-loading complex. 𝞫 subunits make two clamps with a dimer each. They increase the processivity of the DNA polymerase III.
DNA Polymerase IV is coded by dinB gene. Its main role is in DNA repair during SOS response, when DNA replication is stalled at the replication fork. DNA polymerase II, IV and V are translesion polymerases.
DNA Polymerase V is also involved in translesion synthesis during SOS response and DNA repair. It is made up of UmuC monomer and UmuD dimer.
Eukaryotic DNA Polymerase Types and Function
Like prokaryotic cells, eukaryotic cells also have many DNA polymerases, which perform different functions, e.g. mitochondrial DNA replication, nuclear DNA replication, etc. The nuclear DNA replication is mainly done by DNA polymerase 𝝳 and 𝜶. There are at least 15 DNA polymerases identified in human beings.
- DNA polymerase 𝝳 – It is the main enzyme for replication in eukaryotes. It also has 3’→5’ exonuclease activity for proofreading.
- DNA polymerase 𝜶 – The main function of DNA polymerase 𝜶 is to synthesize primers. The smaller subunit has a primase activity. The largest subunit has polymerization activity. It forms a primer for Okazaki fragments, which is then extended by DNA polymerase 𝝳.
- DNA polymerase 𝟄 – The main function is DNA repair. It removes primers for Okazaki fragments from the lagging strand.
- DNA polymerase 𝝲 – It is the main replicative enzyme for mitochondrial DNA.
| Also Check: MCQs on DNA Replication |
How does DNA Polymerase work?
The reaction is phosphoryl group transfer. The 3’-OH group of the growing strand acts as a nucleophile and attacks the incoming deoxyribonucleoside triphosphate at the 𝜶-phosphorus, leading to phosphodiester bond formation. Inorganic phosphate is released in the reaction.
(dNMP)n + dNTP → (dNMP)n+1 + PPi
All the DNA polymerases require two Mg ions at the active site. It is important to note that DNA polymerase can only add nucleotides at the 3′ end of the growing strand, that is why replication always occurs in the 5’→3’ direction. They cannot initiate the formation of new DNA.
They need a template strand, which guides the polymerisation reaction. They also need a primer for their action as they can only add nucleotides at 3’ OH group. The primer can be a short segment of RNA, DNA or both. Generally, the primer is an RNA oligonucleotide in the living system.
After adding a nucleotide, the DNA polymerase can either dissociate or move along to add more nucleotides. It depends on the processivity of DNA polymerase and it differs in different DNA polymerases.
Replication is a highly accurate process and even the change in a single nucleotide can cause mutation. To avoid this there are two mechanisms by which DNA polymerases ensure that there are no discrepancies.
- The geometry of the active sites allows only the correct nucleotide base pairs to fit. But this is not sufficient and it is seen that it can add an incorrect nucleotide after correctly adding 104 to 105 nucleotides.
- To correct this type of errors, DNA polymerase has 3’→5’ exonuclease activity. DNA polymerase checks each of the added nucleotides and removes the nucleotide if there is a mismatch. This is known as proofreading. In DNA polymerase I, there are different active sites for polymerizing and proofreading functions.
This was all about DNA Polymerase. Explore notes on Molecular Basis of Inheritance to know in detail about the replication process, only at BYJU’S.
Frequently Asked Questions
What is the role of DNA polymerase?
The main function of DNA polymerase is to synthesize DNA by the process of replication. It adds deoxyribonucleotides at the 3′-OH group of the growing DNA strand and synthesises the new strand in 5’→3’ direction.
Also see: Nucleotide
Different DNA polymerases perform specific functions. In prokaryotes, DNA polymerase III is the main enzyme responsible for replication. DNA polymerase I and II have a role to play in repair, removing the primer and filling the gaps. In eukaryotes, DNA polymerase 𝝳 is the main enzyme for replication. Other DNA polymerases are involved in the repair, proofreading and primer removal.
What are the 3 main functions of DNA polymerase?
The main function of DNA polymerase is to replicate and form new DNA strands and repair any mismatch or damage in the DNA. DNA polymerase duplicates the cellular DNA content every time a cell divides so that there is an equal distribution of DNA to the daughter cells. The three main functions of DNA polymerase are:
- 5’→3’ polymerisation – it is required for replication and to add nucleotides at the 3’-OH group of the growing DNA strand and filling the gaps.
- 3’→5’ exonuclease – it is required for proofreading and DNA polymerase removes any incorrectly added nucleotides while replication.
- 5’→3’ exonuclease – It is responsible for removing RNA primers and repair.
What are the types of DNA polymerase?
There are various different types of DNA polymerase identified in prokaryotes and eukaryotes:
- Prokaryotes contain five different DNA polymerases named from I to V.
DNA polymerase III – is the main enzyme responsible for replication. Other DNA polymerases take part in repair, removing, primer, proofreading, translesion synthesis. - Eukaryotes also contain many different types of DNA polymerase.
DNA polymerase 𝝳 and 𝜶 – The main DNA polymerases for nuclear replication.
DNA polymerase 𝝲 – It is involved in mitochondrial DNA replication.
DNA polymerase 𝟄 – Its main function is to repair DNA. It removes primer from Okazaki fragments.
Which DNA polymerase is used in DNA replication?
DNA polymerase III is used in the replication process in prokaryotic cells and DNA polymerase 𝝳 is the main enzyme for replication in eukaryotic cells.
What’s the difference between DNA polymerase and RNA polymerase?
DNA polymerase synthesises DNA during replication and RNA polymerase synthesises RNA during transcription.
Does DNA polymerase need a primer?
Yes, DNA polymerase requires a primer as they can add a nucleotide to the 3’-OH group of a DNA strand. DNA polymerases cannot initiate the replication process and they need a primer to add nucleotides.
What is the difference between DNA polymerase 1 and 3?
DNA polymerase 3 is the main enzyme catalysing the 5’→3’ polymerisation of DNA strand during replication. It also has 3’→5’ exonuclease activity for proofreading. Whereas DNA polymerase 1 is the main enzyme for repair, removal of primers and filling the gaps in the lagging strand. Apart from polymerisation and 3’→5’ exonuclease activity like DNA polymerase 3, it also has 5’→3’ exonuclease activity.
Related Articles:
| Important Notes for NEET – Chromosome Structure |
| NEET Biology Flashcards – Molecular Basis of Inheritance |
| NEET Questions – Molecular Basis of Inheritance |

Comments