Flashcards for NEET Biology are designed to boost your NEET preparation. Find below flashcards for Molecular Basis of Inheritance. These flashcards on the Molecular Basis of Inheritance are prepared as per the NEET syllabus. This is helpful for aspirants of NEET and other exams during last-minute revision. Flashcards For NEET Biology – Molecular Basis of Inheritance, covers all the important points that are frequently asked in the exam. Check BYJU’S for the full set of Flashcards and Study material for NEET Biology. Solve NEET Biology MCQs to check your understanding and outperform in the exam.
Download PDF of NEET Biology Flashcards for Molecular Basis of Inheritance
| Name of the NEET sub-section | Topic | Flashcards helpful for |
| Biology | Molecular Basis of Inheritance | NEET exams |
| Molecular Basis of Inheritance | |
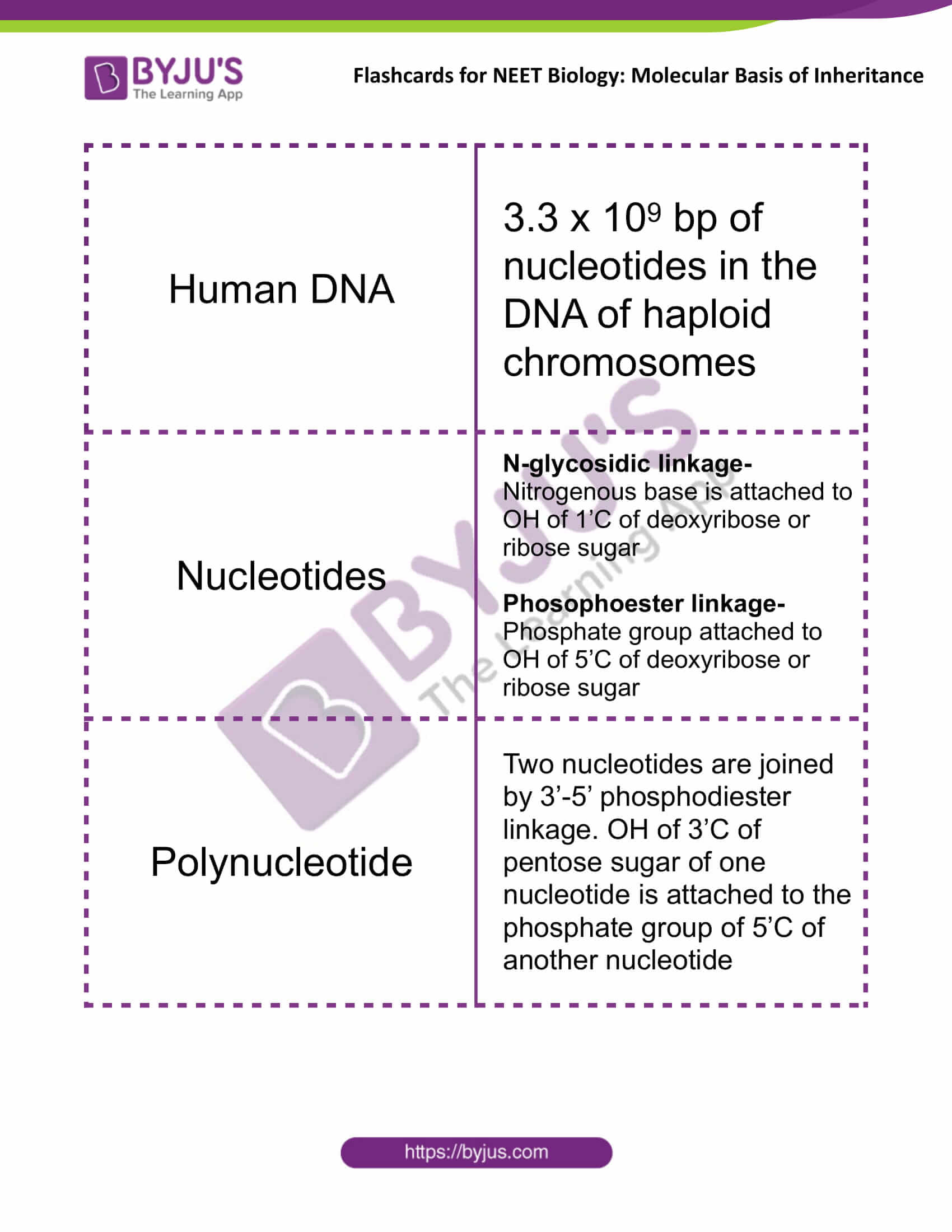
| Human DNA | 3.3 x 109 bp of nucleotides in the DNA of haploid chromosomes |
| Nucleotides | N-glycosidic linkage- Nitrogenous base is attached to OH of 1’C of deoxyribose or ribose sugar
Phosophoester linkage- Phosphate group attached to OH of 5’C of deoxyribose or ribose sugar |
| Polynucleotide | Two nucleotides are joined by 3’-5’ phosphodiester linkage. OH of 3’C of pentose sugar of one nucleotide is attached to the phosphate group of 5’C of another nucleotide |
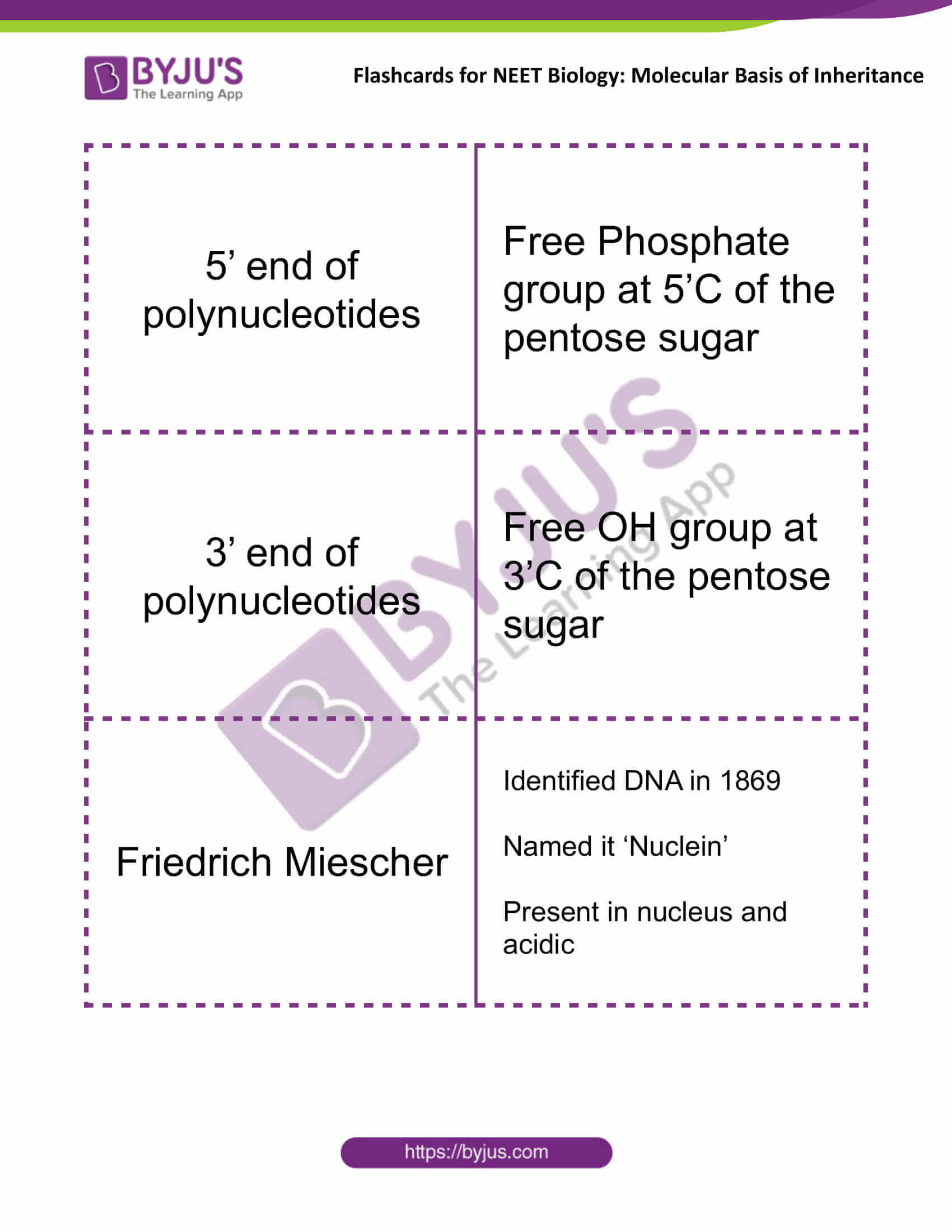
| 5’ end of polynucleotides | Free Phosphate group at 5’C of the pentose sugar |
| 3’ end of polynucleotides | Free OH group at 3’C of the pentose sugar |
| Friedrich Miescher | Identified DNA in 1869
Named it ‘Nuclein’ Present in the nucleus and acidic |
| Maurice Wilkins and Rosalind Franklin | Obtained DNA images from X-ray crystallography |
| James Watson and Francis Crick | Proposed the double-helix model of DNA in 1953 |
| Erwin Chargaff | Elucidated that the DNA contains equal amounts of adenine and thymine and in the same way guanine and cytosine |
| DNA double helix | Antiparallel strands, coiled in a right-handed fashion
Backbone- sugar-phosphate Bases are bonded by hydrogen bonds and projected inside Adenine bonds to thymine by two hydrogen bonds Guanine bonds to cytosine by three hydrogen bonds Pitch- 3.4 nm and 10 bp in each turn |
| Central Dogma | Proposed by Francis Crick in 1958
The flow of genetic information from DNA → RNA → Protein |
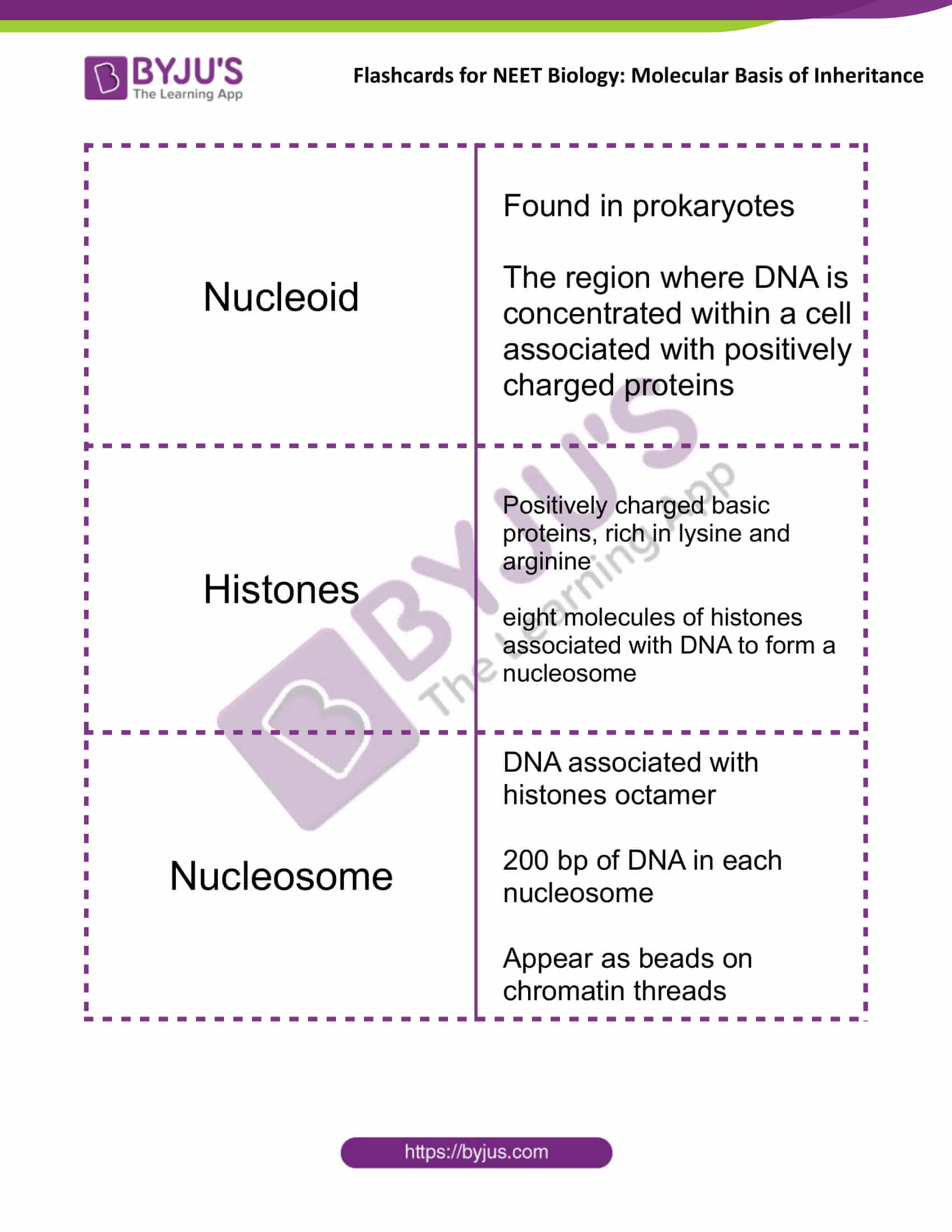
| Nucleoid | Found in prokaryotes
The region where DNA is concentrated within a cell associated with positively charged proteins |
| Histones | Positively charged basic proteins, rich in lysine and arginine
Eight molecules of histones associated with DNA to form a nucleosome |
| Nucleosome | DNA associated with histones octamer
200 bp of DNA in each nucleosome Appear as beads on chromatin threads |
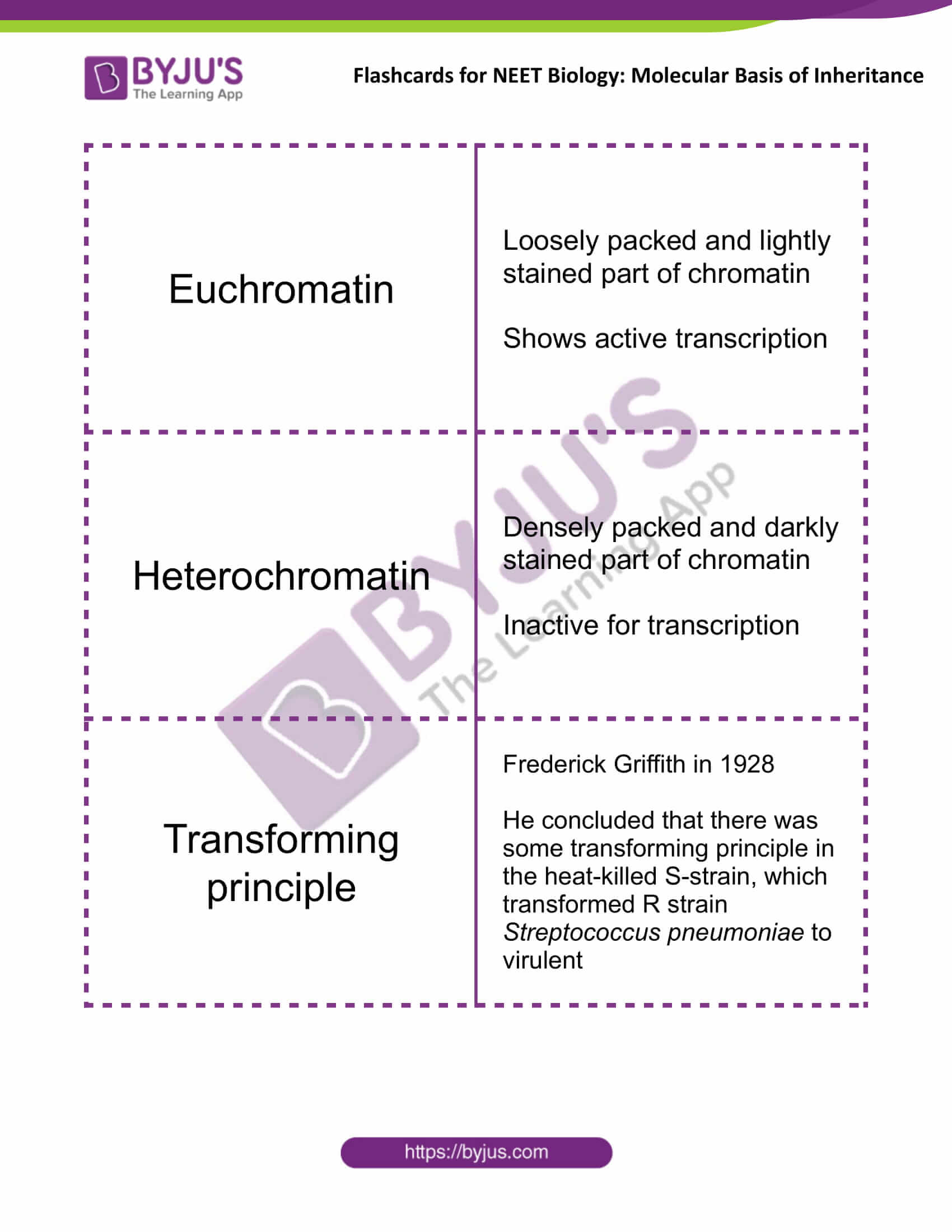
| Euchromatin | Loosely packed and lightly stained part of chromatin
Shows active transcription |
| Heterochromatin | Densely packed and darkly stained part of chromatin
Inactive for transcription |
| Transforming principle | Frederick Griffith in 1928
He concluded that there was some transforming principle in the heat-killed S-strain, which transformed R strain Streptococcus pneumoniae to virulent |
| Avery, MacLeod and McCarty | Demonstrated that the DNA caused the bacterial transformation
They showed that proteases and RNAases did not affect transformation, whereas DNAase inhibit transformation |
| Alfred Hershey and Martha Chase | Unequivocal proof of DNA as genetic material in 1952
They worked on bacteriophages Radioactive 35S labelled- protein capsule Radioactive 32P labelled- DNA |
| RNA as a genetic material | Present in many viruses, known as a retrovirus
E.g. TMV, QB bacteriophage, etc. ds RNA- reovirus, etc. |
| Semiconservative DNA replication | Proposed by Watson and Crick
Experimentally proved by Matthew Meselson and Franklin Stahl in 1958 using 15N isotope in the growing medium for E.coli Taylor and colleagues worked on Vicia faba using radioactive thymidine |
| DNA-dependent DNA polymerase | Main enzyme for replication
Polymerisation in 5’→3’ direction |
| DNA ligase | Join the DNA strand, which is synthesised discontinuously during replication |
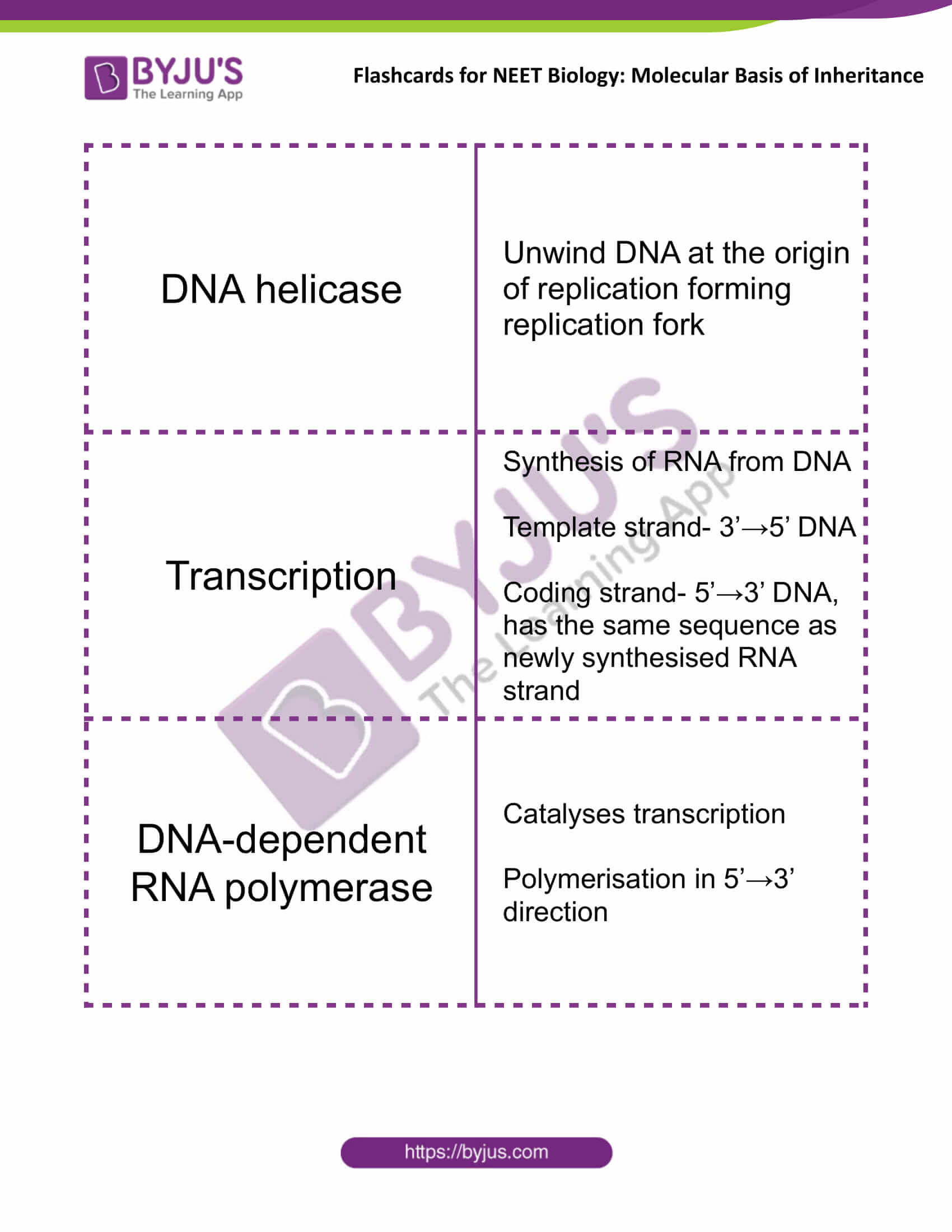
| DNA helicase | Unwind DNA at the origin of replication forming replication fork |
| Transcription | Synthesis of RNA from DNA
Template strand- 3’→5’ DNA Coding strand- 5’→3’ DNA, has the same sequence as newly synthesised RNA strand |
| DNA-dependent RNA polymerase | Catalyses transcription
Polymerisation in 5’→3’ direction |
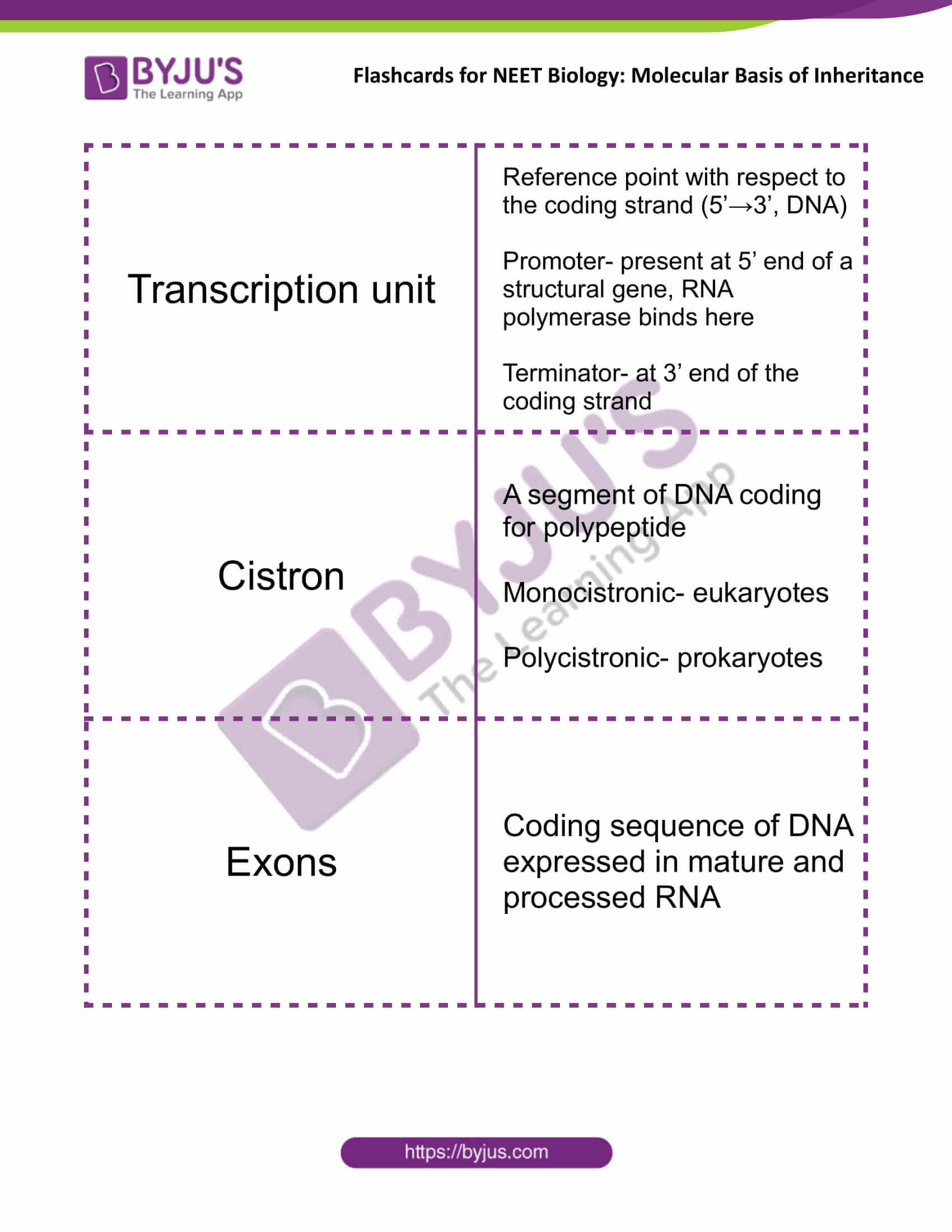
| Transcription unit | Reference point with respect to the coding strand (5’→3’, DNA)
Promoter- present at 5’ end of a structural gene, RNA polymerase binds here Terminator- at 3’ end of the coding strand |
| Cistron | A segment of DNA coding for polypeptide
Monocistronic- eukaryotes Polycistronic- prokaryotes |
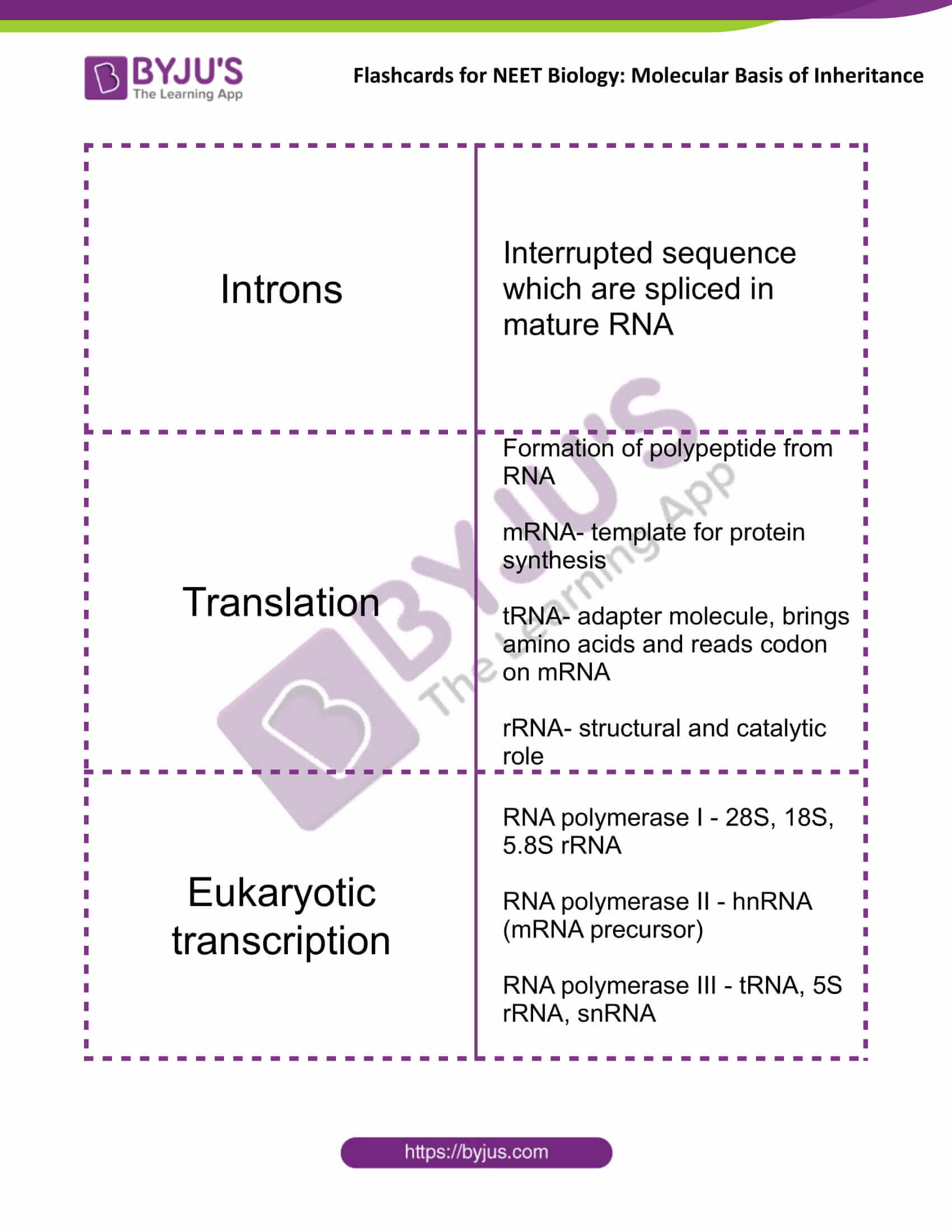
| Exons | Coding sequence of DNA expressed in mature and processed RNA |
| Introns | Interrupted sequence which are spliced in mature RNA |
| Translation | Formation of polypeptide from RNA
mRNA- template for protein synthesis tRNA- adapter molecule, brings amino acids and reads codon on mRNA rRNA- structural and catalytic role |
| Eukaryotic transcription | RNA polymerase I – 28S, 18S, 5.8S rRNA
RNA polymerase II – hnRNA (mRNA precursor) RNA polymerase III – tRNA, 5S rRNA, snRNA |
| hnRNA processing | Splicing – removal of introns and joining together of exons
Capping – addition of methyl guanosine triphosphate at 5’ end Tailing – addition of adenylate residues (200-300) at 3’ end |
| Marshall Nirenberg | Deciphered first the 64 triplet codons for 20 amino acids present in protein |
| Start codon | AUG
Codes for Methionine |
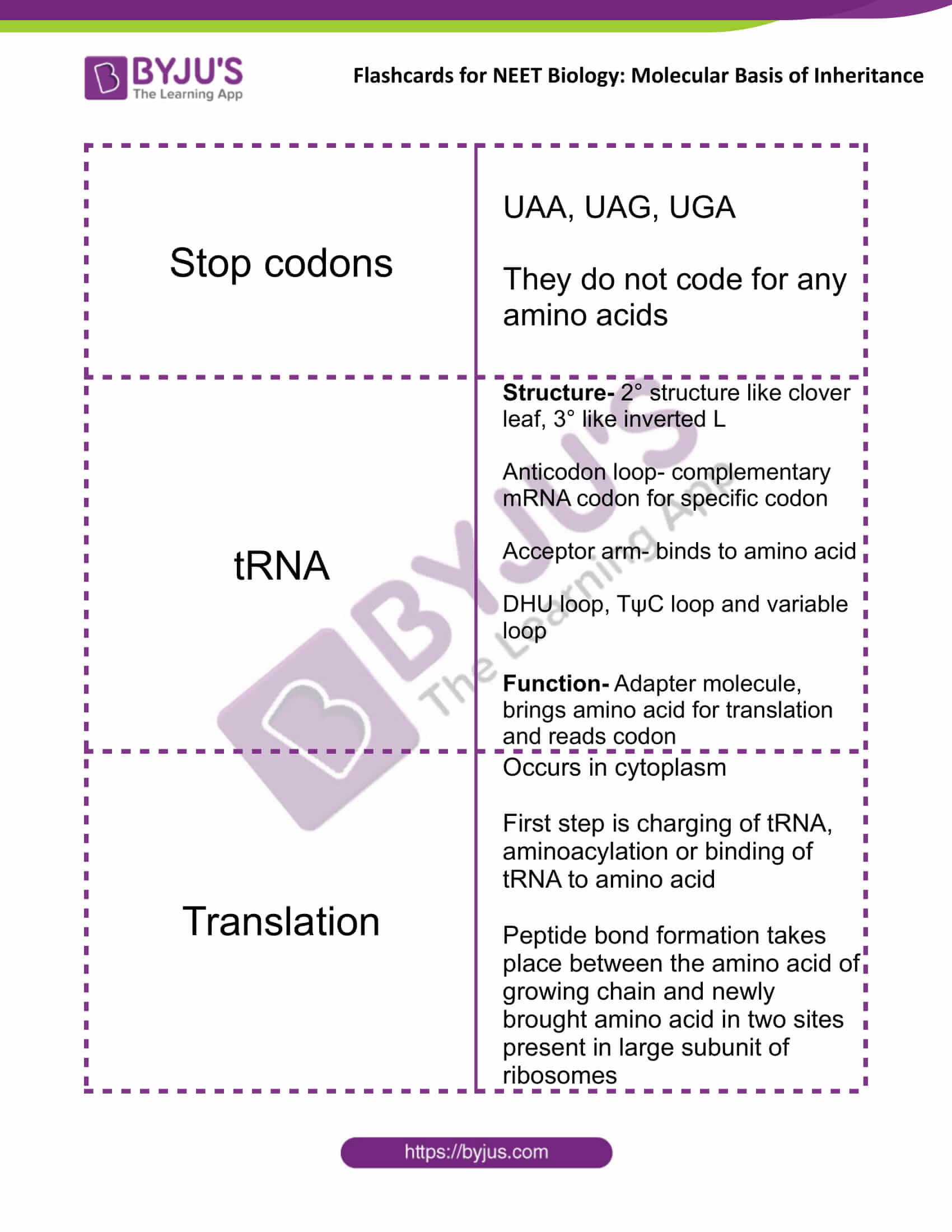
| Stop codons | UAA, UAG, UGA
They do not code for any amino acids |
| tRNA | Structure- 2° structure like clover leaf, 3° like inverted L
Anticodon loop- complementary mRNA codon for specific codon Acceptor arm- binds to amino acid DHU loop, TψC loop and variable loop Function- Adapter molecule, brings amino acid for translation and reads codon |
| Translation | Occurs in cytoplasm
The first step is charging of tRNA, aminoacylation or binding of tRNA to amino acid Peptide bond formation takes place between the amino acid of growing chain and newly brought amino acid in two sites present in the large subunit of ribosomes |
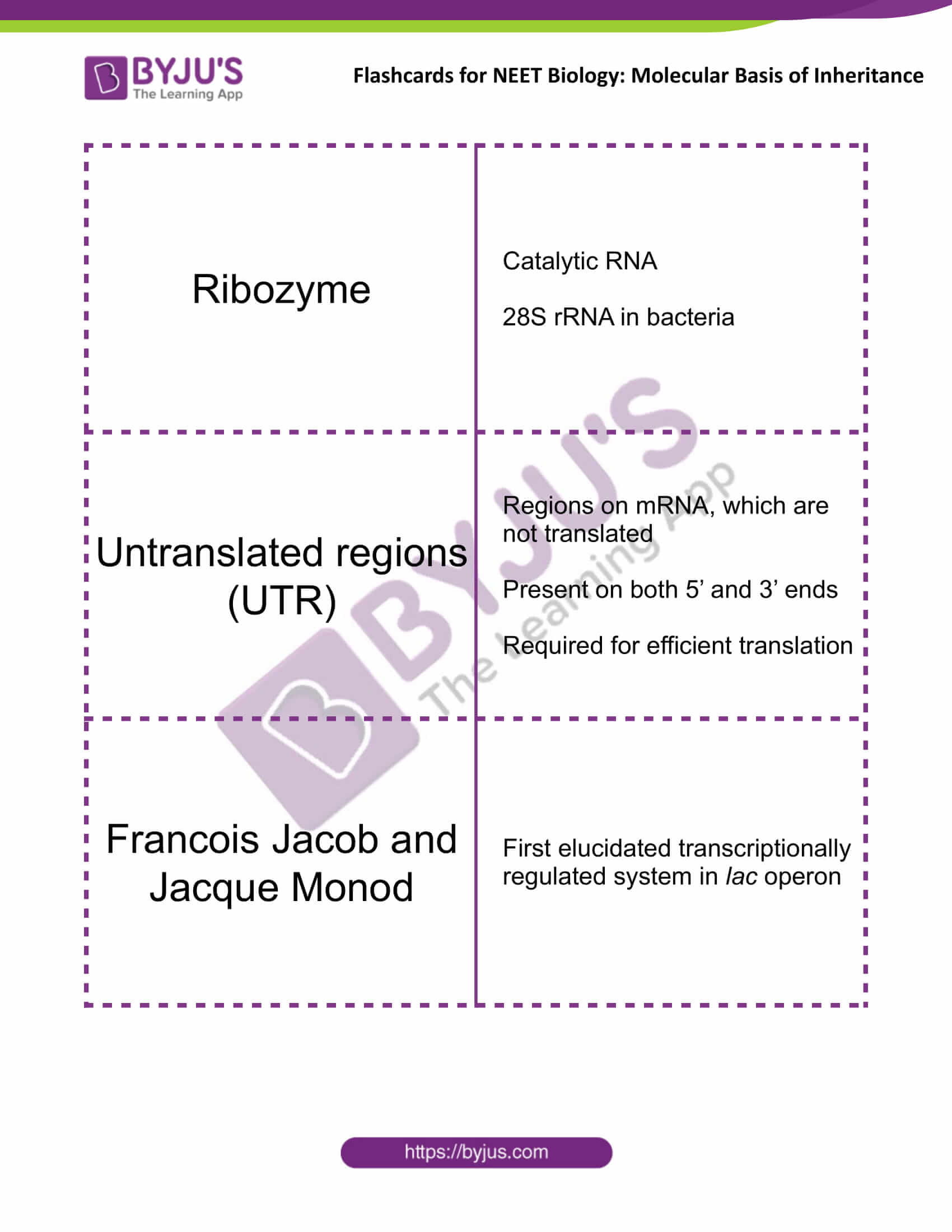
| Ribozyme | Catalytic RNA
28S rRNA in bacteria |
| Untranslated regions (UTR) | Regions on mRNA, which are not translated
Present on both 5’ and 3’ ends Required for efficient translation |
| Francois Jacob and Jacque Monod | First elucidated transcriptionally regulated system in lac operon |
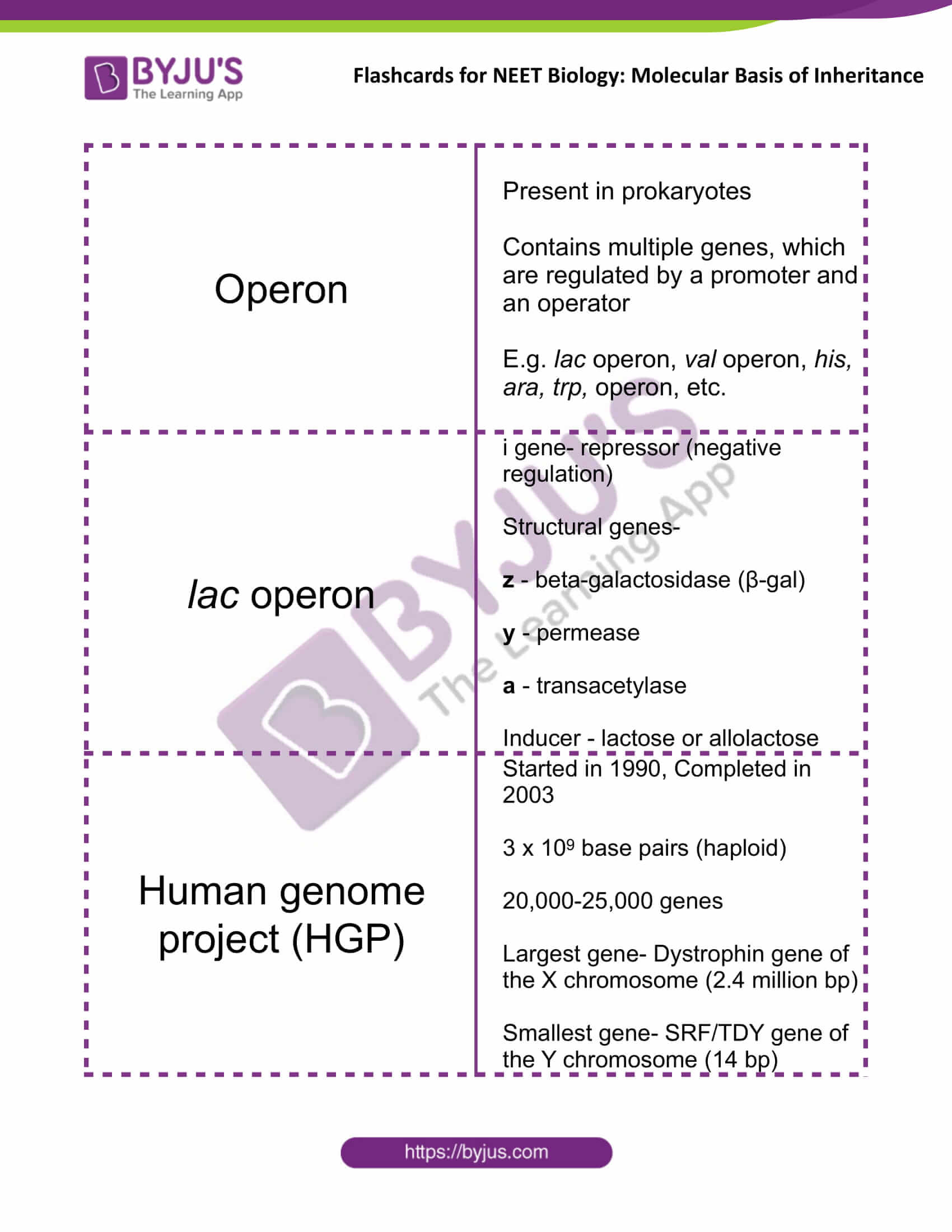
| Operon | Present in prokaryotes
Contains multiple genes, which are regulated by a promoter and an operator E.g. lac operon, val operon, his, ara, trp, operon, etc. |
| lac operon | i gene- repressor (negative regulation)
Structural genes- z – beta-galactosidase (β-gal) y – permease a – transacetylase Inducer – lactose or allolactose |
| Human genome project (HGP) | Started in 1990, Completed in 2003
3 x 109 base pairs (haploid) 20,000-25,000 genes Largest gene- Dystrophin gene of the X chromosome (2.4 million bp) Smallest gene- SRF/TDY gene of the Y chromosome (14 bp) |
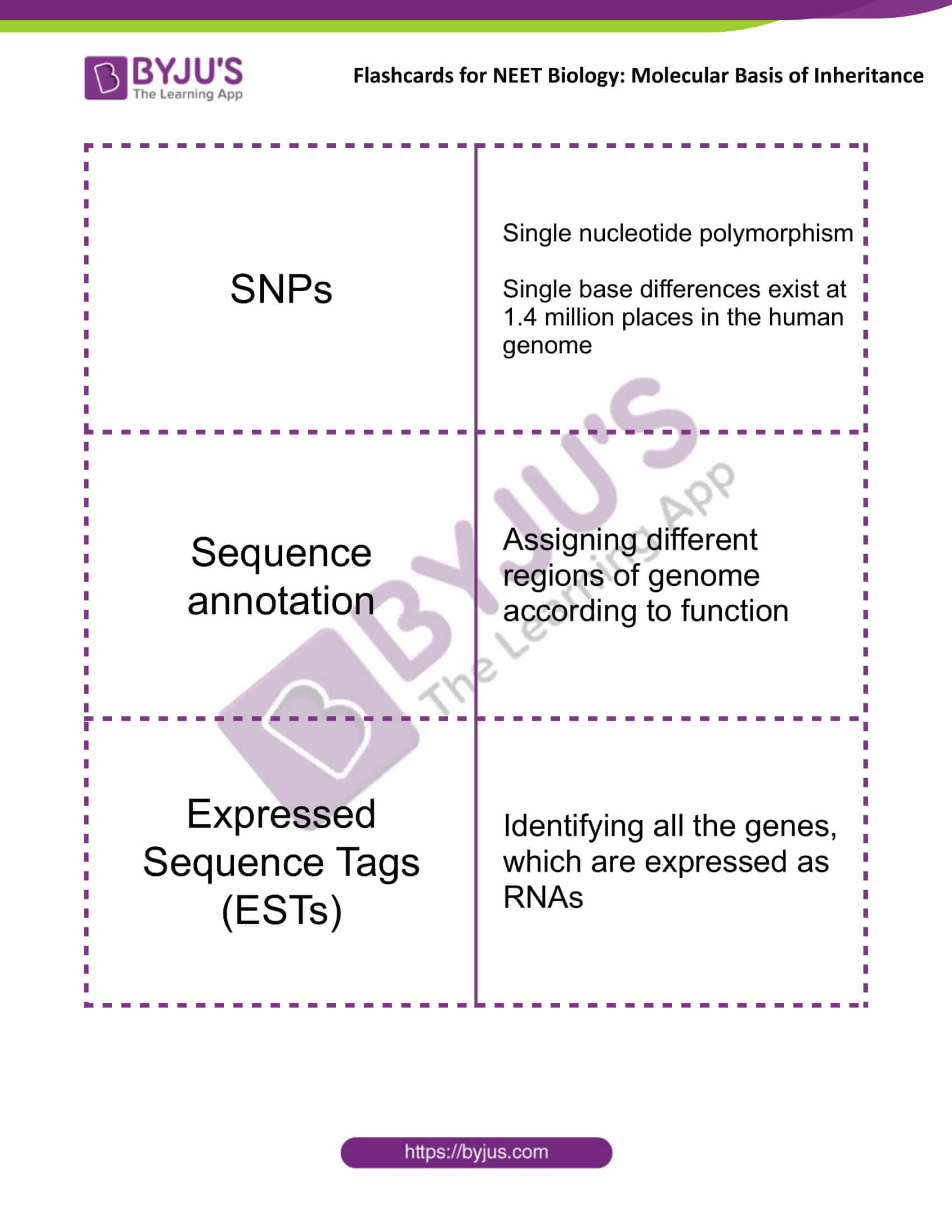
| SNPs | Single nucleotide polymorphism
Single base differences exist at 1.4 million places in the human genome |
| Sequence annotation | Assigning different regions of genome according to function |
| Expressed Sequence Tags (ESTs) | Identifying all the genes, which are expressed as RNAs |
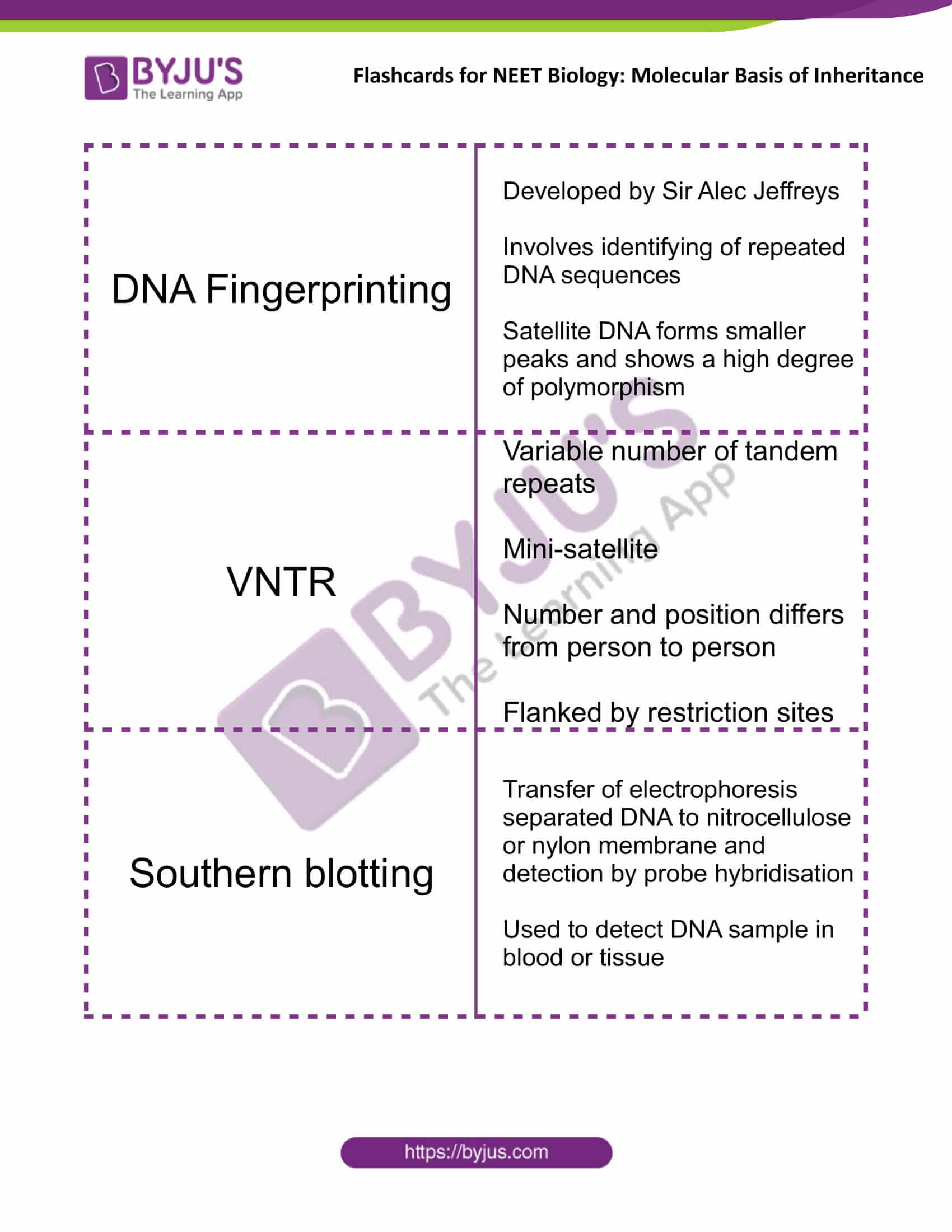
| DNA Fingerprinting | Developed by Sir Alec Jeffreys
Involves identifying of repeated DNA sequences Satellite DNA forms smaller peaks and shows a high degree of polymorphism |
| VNTR | Variable number of tandem repeats
Mini-satellite Number and position differs from person to person Flanked by restriction sites |
| Southern blotting | Transfer of electrophoresis separated DNA to nitrocellulose or nylon membrane and detection by probe hybridisation
Used to detect DNA sample in blood or tissue |
Get access to the full set of flashcards for NEET Biology, only at BYJU’S.
Recommended Video:
Also Check:
NEET Flashcards: Human Reproduction
NEET Flashcards: Principles Of Inheritance And Variation





Very nice mam