Find below the important notes for the chapter, Molecular Basis of Inheritance as per NEET Biology syllabus. This is helpful for aspirants of NEET and other exams during last-minute revision. Important notes for NEET Biology- Molecular Basis of Inheritance covers all the important topics and concepts useful for the exam. Check BYJU’S for the full set of important notes and study material for NEET Biology and solve the NEET Biology MCQs to check your understanding of the subject.
Download Complete Chapter Notes of Molecular Basis of Inheritance
Download Now
| Name of the NEET sub-section | Topic | Notes helpful for |
| Biology | Molecular Basis of Inheritance | NEET exams |
Also see: NEET Answer Key
Molecular Basis of Inheritance – Important Points, Summary, Revision, Highlights
DNAPolynucleotide chainDouble Helix ModelPackaging of DNAReplicationTranscriptionGenetic CodeMutationTranslationCentral DogmaRegulationLac OperonHGPDNA Fingerprinting
The DNA (Deoxyribonucleic acid)
- DNA is the genetic material in most of the organisms except some viruses, which have an RNA genome, e.g. TMV (Tobacco mosaic virus)
- RNA mostly acts as a messenger, an adapter and has a catalytic function
- Number of base pairs (bp) or nucleotides determines the length of the DNA
Human DNA (haploid) – 3.3 x 109 bp
Bacteriophage 𝜙 x 174 – 5386 nucleotides
Bacteriophage 𝝀 – 48502 bp
E. coli – 4.6 x 106 bp
- Friedrich Meischer in 1869 identified DNA present in the nucleus as an acidic substance and named it as ‘Nuclein’
- Frederick Griffith in 1928 demonstrated in the infected mice that some “transforming principle” gets transferred from heat-killed S-strain of Streptococcus pneumoniae to R-strain, enabling it to make the smooth polysaccharide coat, hence it becomes virulent
- Oswald Avery, Colin MacLeod and Maclyn McCarty determined the biochemical nature of “transforming principle” and discovered that only DNA is responsible for the transformation
- Hershey and Chase in 1952 proved that DNA is the genetic material. They infected the bacteria E. coli with bacteriophages grown in radioactive phosphorus (32P) which labels DNA of the bacteriophage, which gets transferred to the bacteria cells and radioactive sulfur (35S), which labels the protein coat of the bacteriophage, hence radioactivity is not detected in the bacterial cell
Structure of a polynucleotide chain
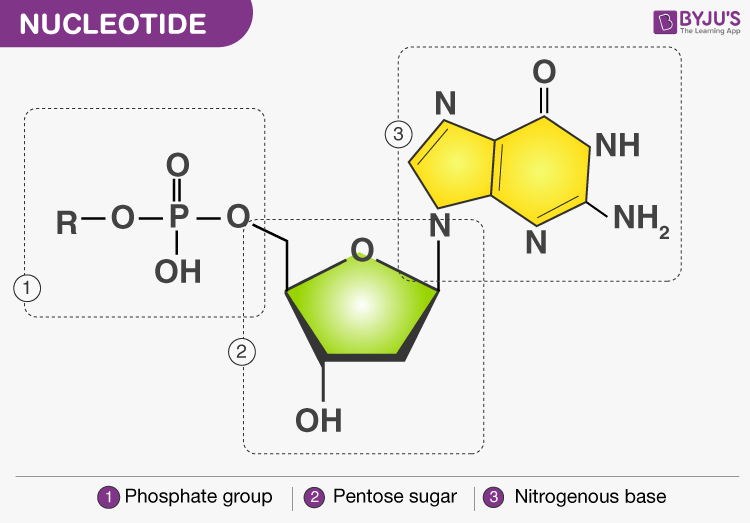
- Each nucleotide is composed of three elements:
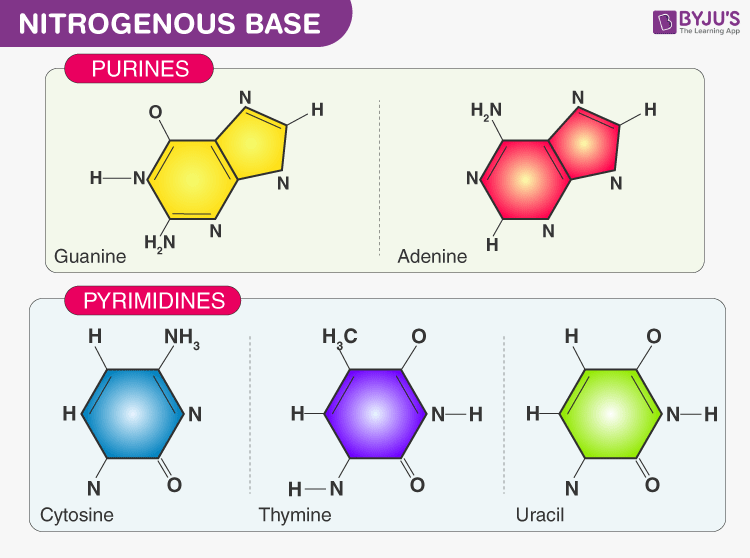
- Nitrogenous base:
- Purines- Adenine (A) and Guanine (G) present in DNA as well as RNA
- Pyrimidines- Cytosine (C) and Thymine (T) in DNA and Cytosine and Uracil in RNA. Thymine is also known as 5-methyl uracil and it is accounted for more stability of DNA molecule
- Sugar: Pentose sugar- Ribose in RNA (ribonucleic acid), deoxyribose in DNA
- Phosphate group
- Nitrogenous base:
- Nucleoside: a nitrogenous base linked to the hydroxyl group of 1’ C of the pentose sugar by N-glycosidic bond
- Nucleotide: phosphate group is attached to the hydroxyl group present at 5’ C of the nucleoside by a phospho-ester bond
- 3’-5’ phosphodiester bond links two nucleotides together to form dinucleotide and the chain continues to grow to form a polynucleotide
Double Helix Model for Structure of DNA
- Watson and Crick in 1953 proposed the double helix structure of DNA
- Ervin Chargaff observed that the ratio of Adenine and Thymine to Guanine and Cytosine is one and remains constant
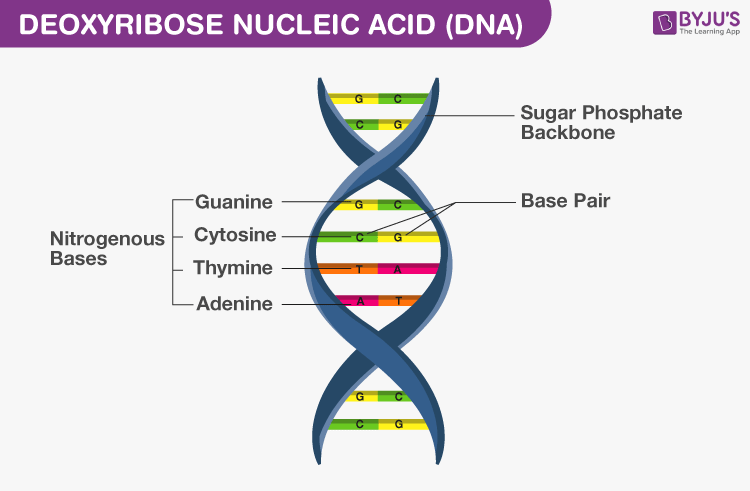
- Two polynucleotide chains make DNA, where the backbone is of sugar-phosphate and bases are present towards inside
- The two chains have the opposite polarity, i.e. one with 5’→3’ and the other has 3’→5’ polarity
- Base pair (bp) is formed by hydrogen bonding between nitrogenous bases of both the polypeptide chains
- Always a purine base of one nucleotide chain is linked to a pyrimidine base of another nucleotide chain or vice versa to make a base pair
- Adenine pairs with Thymine (or Uracil in RNA) by two hydrogen bonds (A=T)
- Guanine pairs with Cytosine by three hydrogen bonds (G)
- The two polypeptide chains are coiled in a right-handed direction
- The distance between two base pairs is 0.34 nm, there are 10 bp in each turn and the pitch of the helix id 3.4 nm
- Stability of the helical structure is conferred by the presence of base pairs one above the other
Packaging of DNA Helix
- In prokaryotes, DNA is organised as a large loop in the nucleoid region. Negatively charged DNA is held together by positively charged proteins
- In eukaryotes there occurs a complex organisation of DNA in chromosomes. DNA is wound around the core of histone octamer (a unit with 8 histone molecules) to form a nucleosome
- Histones are positively charged proteins as they are rich in basic amino acids; lysine and arginine
- There are 5 types of histone proteins- H1, H2A, H2B, H3 and H4
- Histone octamer has 2 molecules of 4 histone proteins and it plays an important role in gene regulation
- A nucleosome is a repeating unit in chromatin, it prevents DNA from getting tangled
- Nucleosome contains around 200 bp of DNA
- Non-histone chromosomal proteins (NHC) help in further packaging of chromatin
- Euchromatin: these are transcriptionally active areas, where chromatin is loosely packed and they take up light stain
- Heterochromatin: these are transcriptionally inactive areas, where chromatin is densely packed and they take up dark stain
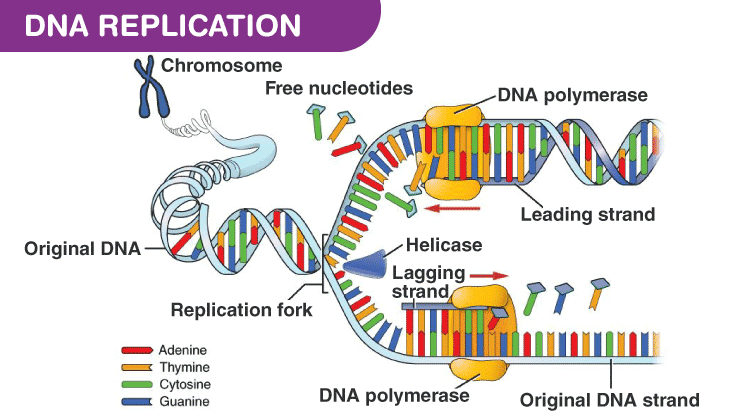
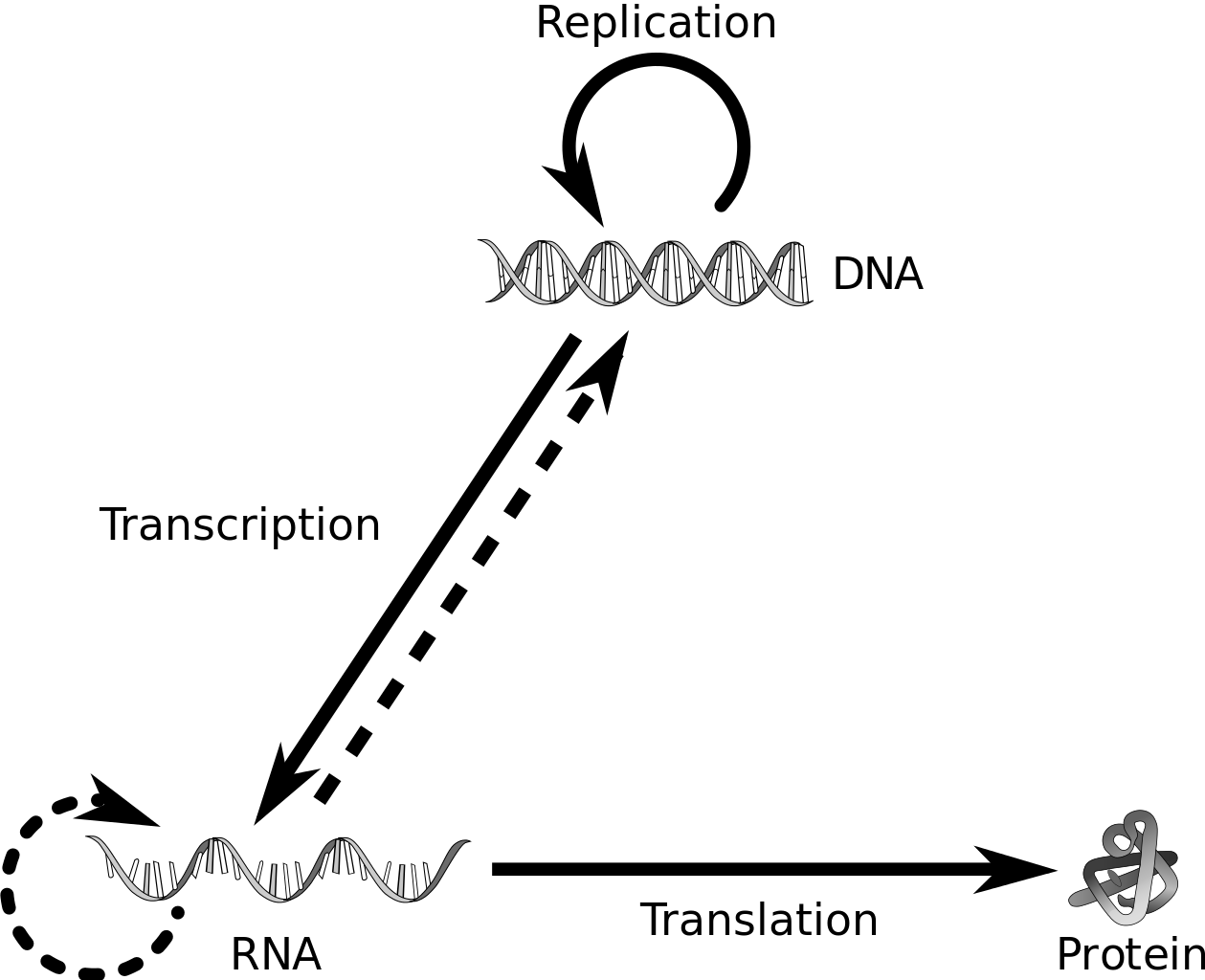
Replication
- Watson and Crick suggested that the replication of DNA is semiconservative
- Meselson and Stahl in 1958 experimentally proved that the DNA replicates semi conservatively
- Taylor et al in another experiment on fava beans (Vicia faba) using radioactive thymidine proved that the replication on DNA is semiconservative
- Enzyme DNA polymerase catalyses DNA replication. It can polymerise only in 5’→3’ direction
- Replication is initiated at the origin of replication
- Deoxyribonucleoside triphosphate provides energy for the polymerisation reaction and also acts as a substrate
- A small part of DNA opens up making a replication fork, where replication occurs
- Replication is continuous in a strand with 5’→3’ direction, called leading strand, where the template strand has 3’→5’ polarity, called leading strand template
- Replication is discontinuous in the other strand, where the template strand has 5’→3’ polarity, called lagging strand template
- The discontinuous fragments, called Okazaki fragments are joined together by the enzyme DNA ligase
- In eukaryotic cells, the replication takes place during s-phase of the cell cycle
- If cell division doesn’t occur after the replication, it results in polyploidy of chromosomes
Transcription
- In the process of transcription, the genetic information present in the DNA gets copied to RNA
- Only one segment of DNA gets copied to RNA
- In RNA Uracil is present instead of Thymine in DNA
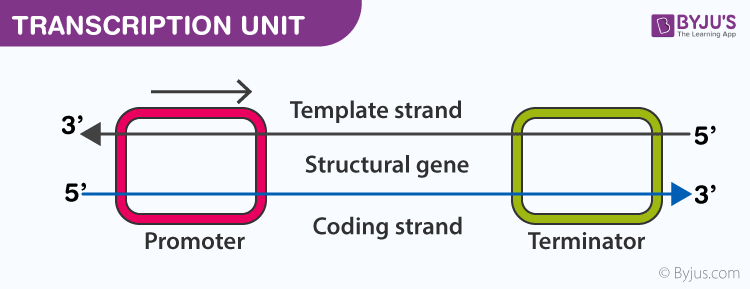
- Transcription of DNA includes three regions; a promoter, the structural gene and a terminator
- RNA polymerase catalyses the transcription and direction of transcription is same as that of replication by DNA polymerase, i.e. 5’→3’ direction
- Template Strand: it has 3’→5’ polarity, which acts as a template for RNA formation, also called antisense strand
- Coding strand: it has 5’→3’ polarity, which has the sequence similar to the newly formed RNA, except that Thymine is replaced by Uracil in RNA, also called the sense strand
- Promoter: it is situated at the 5’ side of the structural gene or upstream (with respect to coding strand). RNA polymerase binds here to start the transcription
- Structural gene: the region between promoter and terminator. Cistron is a segment of DNA that codes for a polypeptide. The structural gene is monocistronic in eukaryotic cells and polycistronic in prokaryotic cells
- Terminator: it is situated at the 3’ side of the coding strand and marks the end of the transcription process
- The monocistronic structural gene (in eukaryotes) has interrupted coding sequences, the gene in eukaryotes is called split genes
- Exons: coding sequences, which appear in mature and processed RNA
- Introns: intervening sequences. It is not present in the mature and processed RNA
Transcription in a Prokaryotic cell
- In bacteria, a single DNA-dependent RNA polymerase catalyses the transcription of all the three RNAs present; mRNA, tRNA, rRNA
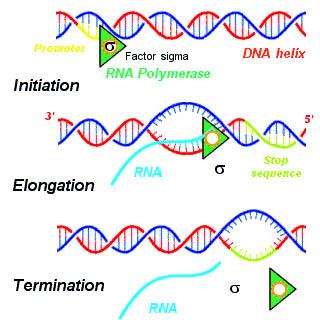
- There are three steps of transcription:
- Initiation: RNA polymerase binds to initiation factor, sigma (𝞂) at the promoter site to start the process of transcription
- Elongation: RNA polymerase is only capable of elongation
- Termination: when the RNA polymerase reaches the terminator region, it binds with the terminator factor, rho (𝜌) to terminate the process and the nascent RNA falls off
- In bacteria, mRNA doesn’t require further processing and as the nucleus and cytosol are not separate, translation is coupled with transcription and is initiated before the full transcription of mRNA occurs
Transcription in a Eukaryotic cell
- There are three RNA polymerases to catalyse transcription of different RNAs in a eukaryotic cell:
- RNA polymerase I- transcription of rRNA (28S, 18S, 5.8S)
- RNA polymerase II- transcription of hnRNA (heterogeneous nuclear RNA), which is a precursor of mRNA
- RNA polymerase III- transcription of tRNA, 5S rRNA and snRNA (small nuclear RNA)
- The primary transcript is non-functional and has exons intervened by introns
- Splicing- the process of removing introns and joining together exons in a defined sequence
- Capping and tailing- additional processing of hnRNA. In capping a methyl guanosine triphosphate is added to the 5’ end of hnRNA. In tailing, 200-300 adenylate residues are added at 3’ end.
- mRNA is fully processed hnRNA, which gets transported out of the nucleus for translation
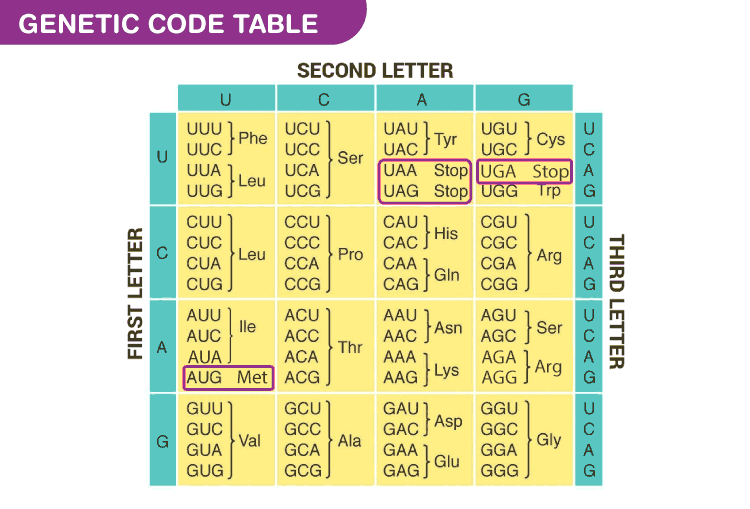
Genetic Code
- It is the sequence of bases in mRNA, that codes for a particular amino acid in the protein synthesis
- Each code is made up of three nucleotides called a triplet. codons are nearly universal, except for some protozoans and mitochondrial codons
- More than one triplet codon code for same amino acid, so the code is degenerate
- There are a total of 64 codons, of which 61 code for amino acids
- 3 codons do not code for any amino acids, they are called stop codons- UAA, UAG, UGA
- AUG is the start codon as well as codes for the amino acid methionine
Genetic Mutation
- Point Mutation: Change of single base pair results in the point mutation, e.g. Sickle cell anaemia is a result of a point mutation in the gene coding for the 𝛽-globin chain. As a result Glutamate in the normal protein gets converted to Valine in the sickle cell
- Frameshift Mutation: When there is loss or gain of one or two base pairs, it changes the reading frame at the point of insertion or deletion resulting in the frameshift mutation
Translation
- The translation is the process of amino acid polymerisation. Amino acids are joined by peptide bonds
- All three RNAs have a different role in the process of translation
- mRNA- provides the template. The sequence of amino acids in a polypeptide chain is determined by the sequence of bases present in mRNA
- tRNA- acts as an adapter, it brings amino acids and reads the genetic code
- rRNA- performs a structural and catalytic role
- tRNA – The Adapter Molecule: Crick proposed the presence of an adapter molecule, which binds to a specific amino acid. It was known as soluble RNA or sRNA and later named as tRNA
>> Shape of the tRNA is like an inverted ‘L’
>> tRNA is specific for each amino acid, there is a specific initiator tRNA
>> There are no tRNAs for stop codons
>> It has an anticodon loop, which has complimentary code present on mRNA
>> There is an amino acid acceptor arm, which binds the specific amino acid as per the codon
- The first step in the process of translation is aminoacylation of tRNA (charging of tRNA)
- Ribosomes are a protein manufacturing factory
- mRNA to protein translation begins with the presence of mRNA in the small subunit of ribosomes
- The process of translation is always in 5’→3’ direction
- Peptide bond formation occurs between two amino acids present on tRNAs in close vicinity
- There are two sites in the large subunit of a ribosome which accommodates two tRNAs with amino acids close enough to form a peptide bond
- Ribosome also acts as a catalyst in the formation of a peptide bond
- A ribozyme is a 23s rRNA molecule present in bacteria, which acts as an enzyme catalysing peptide bond formation
- Start codon and stop codon flank the coding sequence for a polypeptide in mRNA
- Untranslated regions (UTRs)- UTRs are present before the start codon, i.e. at 5’ end and after the stop codon, i.e. towards 3’ end. They are not translated but they make the translation process efficient
- The Release factor binds to the stop codon at the end terminating the process. The polypeptide gets released from the ribosome
Also Check: Difference Between CDS and ORF
The Central Dogma
Francis Crick proposed that genetic information flows from DNA → RNA → Protein, it is known as the central dogma of molecular biology
Regulation of Gene Expression
- Expression of a gene to form polypeptide can be regulated at various levels in eukaryotes
- At the time of formation of a primary transcript, i.e. transcription
- At the time of processing or splicing
- At the time of transportation of mRNA from the nucleus to the cytosol
- At the time of protein synthesis, i.e. translation
- Environmental, physiological and metabolic conditions regulate the gene expression
- The development and differentiation of embryo is a result of coordinated regulation and expression of several sets of genes
- In prokaryotes, control of gene expression is mainly at the initiation of transcription
- The activity of RNA polymerase at the start site is regulated by regulatory proteins, which can be a repressor or activator
- The accessibility of the promoter region is regulated by an operator sequence adjacent to it, that binds with the specific protein, mostly a repressor
- In each operon, there is a specific operator and repressor protein
The Lac Operon
- Jacob and Monad first showed the transcriptionally regulated system in the lac operon
- An operon consists of many structural genes regulated by a common promoter and regulatory gene
- The lac operon consists of
- Regulatory gene: gene i (inhibitor gene), that codes for the repressor of the lac operon
- Structural gene: three structural genes, z, y, a
>> z gene, codes for 𝜷-galactosidase, it hydrolyses lactose to glucose and galactose
>> y gene, codes for permease, responsible for increasing permeability of the cell to 𝛽-galactosides
>> a gene, codes for transacetylase
- The repressor is synthesised continuously from the gene i, which binds to the operator and prevents RNA polymerase from transcribing
- Lactose is a substrate of 𝜷-galactosidase and also regulates the gene expression. It acts as an inducer
- When lactose or allolactose is present, it binds with repressor and inactivates it, allowing RNA polymerase to access to the promoter region and initiating the transcription
- Regulation by repressor in called negative regulation
Human Genome Project
- Human genome project (HGP) was launched in 1990 to decipher the complete DNA sequence of the human genome
- Genetic engineering techniques were used to isolate and clone the DNA segment for determining the DNA sequence
- The project got completed in 2003, the sequence of chromosome 1 was completed in May 2006
- Some of the key findings of HGP are:
>> The human genome has 3164.7 million bp
>> Total ~30,000 genes are present with an average of 3000 bases per gene
>> Dystrophin gene is the largest human gene having 2.4 million bases
>> 99.9 % of nucleotides are the same in all people
>> Only 2 % of genome codes for proteins
>> Most genes are found on chromosome 1, i.e. 2968
>> Least genes are found on the Y chromosome, i.e. 231
>> There are around 1.4 million locations, where there is a single base difference in DNA, it is called single nucleotide polymorphism- SNPs (snips)
DNA Fingerprinting
- Alec Jeffreys initially developed the DNA fingerprinting techniques and named it as VNTR (Variable Number of Tandem Repeats)
- The difference in the DNA makeup accounts for the unique phenotype of each individual and that is the basis of DNA fingerprinting
- The DNA sequence of two individuals can be compared very quickly by the DNA fingerprinting technique
- DNA fingerprinting involves identifying the difference between two DNA molecule at the specific regions where the sequence is repeated many times called repetitive DNA
- These repetitive DNAs make a small peak during density gradient centrifugation and known as satellite DNA
- Depending on the number of repetitive units, base composition and length of the segment, satellite DNA is further classified into micro-satellites, mini-satellites, etc.
- These sequences do not code for protein but make a large portion of the human genome
- A high degree of polymorphism present in these sequences is the basis of DNA fingerprinting
- DNA fingerprinting is used for paternity test as this polymorphism is inherited to the child
- It has been widely used in forensic science
- DNA fingerprinting can be used in determining genetic diversity existing in a population
Explore the next chapter for important points with regards to the NEET exam only at BYJU’S. Check the NEET Study Material for all the important concepts and related topics.
Also see:
Flashcards Of Biology For NEET Molecular Basis Of Inheritance
NEET Flashcards: Principles Of Inheritance And Variation
All my doubts are clear with these notes and thank you Byju’s you helped me out to score best in NEET
LOVE IT
Thank you Byjus these notes are very helpful to me
Hi l am. Muzzamil l am. Study class 12th l am from jummu l want to download pdf of some subject kindly grant give us link
Chapter wise
Sub biology your faithfully
And your abedently
Yo